Postdoctoral fellow
PARCC - Inserm U970
Paris Institute for Transplantation and Organ Regeneration (PITOR)
Application of a novel spatial transcriptomic 6000-plex panel in pig-to-human xenotransplantation
Valentin Goutaudier1, Claire Williams2, Erwan Morgand1, Jeffrey Stern3, Fariza Mezine1, Karen Khalil3, Aurélie Sannier1, Sébastien Giraud4, Thierry Hauet4, Aprajita Mattoo3, Vasishta Tatapudi3, David Ayares5, Patrick Bruneval1, Adam Griesemer3, Massimo Mangiola3, Robert A. Montgomery3, Alexandre Loupy1.
1Paris Institute for Transplantation and Organ Regeneration (PITOR), PARCC - Inserm U970, Paris, France; 2Spatial Biology Department, Bruker Corporation, Billerica, MA, United States; 3NYU Langone Transplant Institute, New York University, New York, NY, United States; 4IRMETIST, Poitiers University, Poitiers, France; 5Research Triangle Park, United Therapeutics Corporation, Durham, NC, United States
Introduction: Kidney xenotransplantation is a promising approach that has the potential to open new treatment avenues for patients currently on transplant waiting lists. Here we applied for the first time a 6000-plex gene panel to decipher the immune response after transplantation of a genetically edited porcine thymokidney into a human recipient.
Methods: We performed in-depth multimodal phenotyping of tissue biopsies taken at 7 sequential follow-up times from Day 0 through Day 61. Initial bulk transcriptomic studies showed a molecular signature evocative of antibody-mediated rejection developing as early as day 10. In addition to a pre-transplant pig kidney control sample, we selected 4 post-transplantation time points to further explore and used the CosMx Spatial Molecular Imager to localize and profile the human immune cells infiltrating the xenograft in their spatial context. We designed a novel bioinformatic approach to accurately distinguish the approximately 3,000 human immune cells from the approximately 70,000 pig structural cells.
Results: Across all time points, macrophages were the most prevalent human cell type, in agreement with previous analyses. The human immune rejection response peaked at the D33 time point after which anti-rejection treatment was provided, leading to a successful decrease in the immune-molecular signs of xenoantibody-mediated rejection. We observed human myeloid cells present in every one of the post-transplant glomeruli in the sampled biopsies and characterized differential expression of pro- and anti-inflammatory myeloid genes over the course of treatment (Figure). We identified spatially-enriched human macrophage – pig structural ligand-receptor interactions present at early and late timepoints and catalogued spatially correlated modules of co-expressed genes.
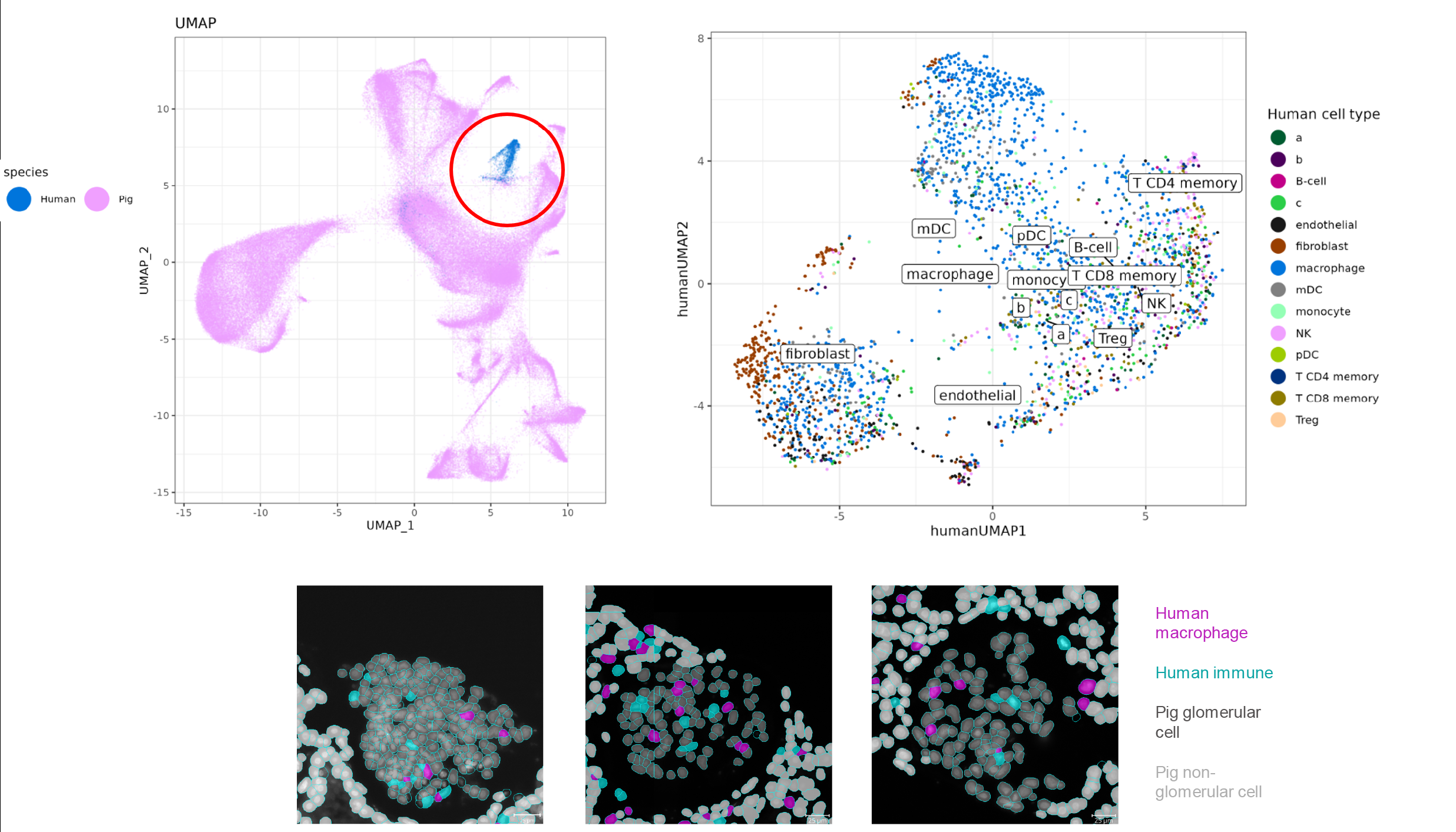
Conclusion: Overall, our data suggest xenoantibody-mediated rejection, characterized by human innate immune cells infiltration in the porcine xenograft, localized within and outside of glomeruli. Discovery of these molecular mechanisms and the successful attenuation of the xenograft organ rejection with targeted treatment paves the way for potential new anti-rejection therapies to further enable human clinical trials.
References:
[1] Spatial Transcriptomics
[2] Precision Medicine
[3] Innate Immunity
[4] Gene Panel
[5] Xenoimmune Response
[6] Therapeutics
Lectures by Valentin Goutaudier
| When | Session | Talk Title | Room |
|---|---|---|---|
|
Thu-02 15:05 - 15:55 |
Innate Immunity and Inflammation 1 | Spatial multimodal characterization of long-term immune response after pig-to-human kidney xenotransplantation | H8-01-F |
|
Thu-02 16:20 - 17:05 |
Innate Immunity and Inflammation 2 | Application of a novel spatial transcriptomic 6000-plex panel in pig-to-human xenotransplantation | H8-01-F |
