
Cell-free DNA method for monitoring porcine-to-human xenotransplants
Tudor Constantin1, Shou-Chien Kuo1, Bernhard Zimmermann1, Hossein Tabriziani1, Philippe Gauthier1.
1Medical affairs, Natera, Inc., Austin, TX, United States
Introduction: Xenotransplantation, the transplantation of organs of non-human origin into humans, is a potential long term solution to the chronic shortage of organs currently available for transplantation. Several xenotransplants using porcine organs from pigs that were genetically modified to remove cell-surface antigens that could lead to alloimmunity, have recently been reported, including heart and kidney, with human patients surviving the surgery for an encouraging duration. Donor-derived cell-free DNA (dd-cfDNA) has been validated as a non-invasive biomarker for the detection of rejection in solid organ transplant, and is routinely used in both a surveillance and for cause setting. A non-invasive cell-free DNA (cfDNA) method for monitoring xenotransplants is needed in anticipation of a large clinical uptake of this approach. In this proof-of-concept study, we sought to demonstrate that cfDNA can be used to measure the fraction of porcine cfDNA (pcfDNA) in plasma.
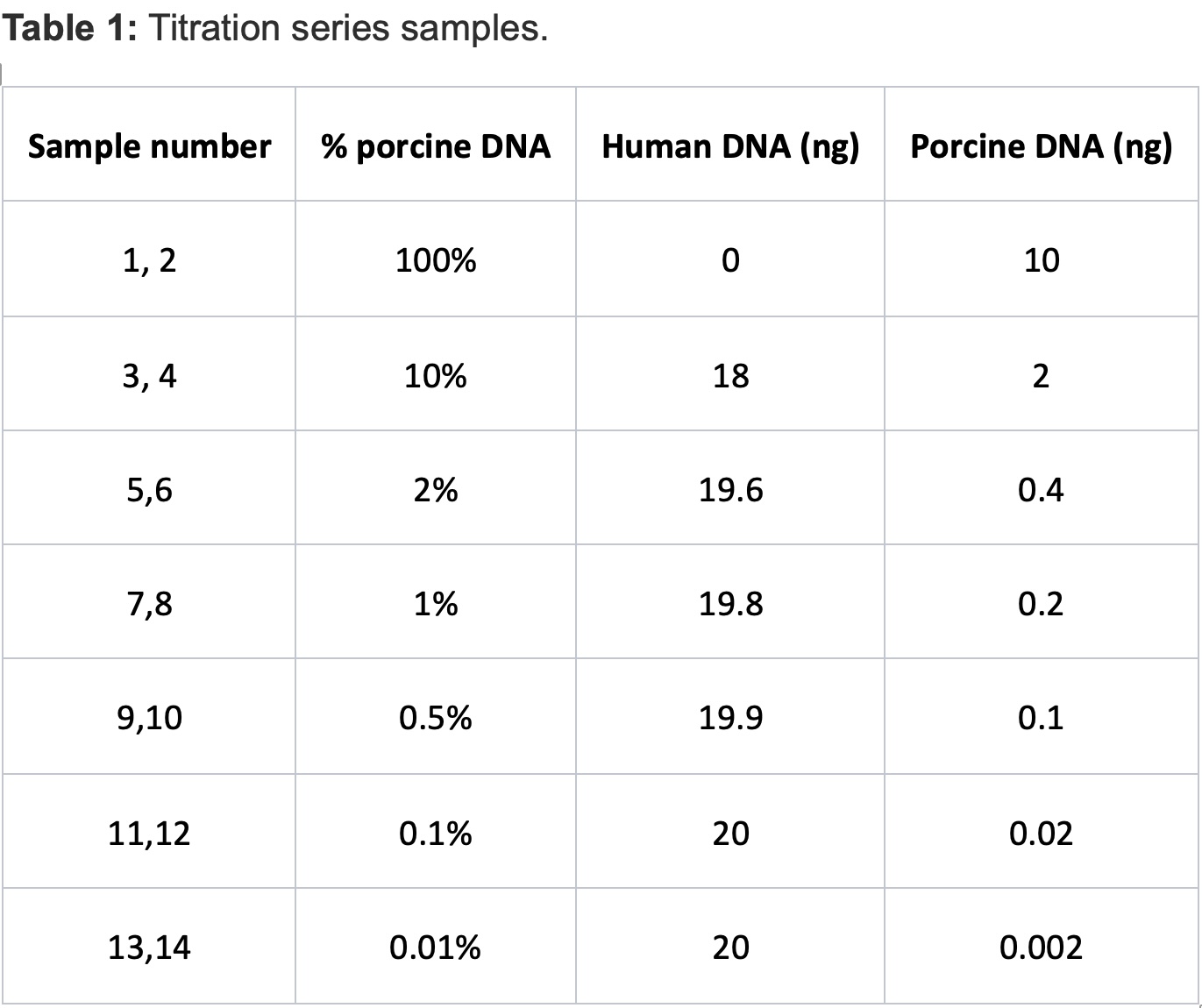
Methods: A titration series of porcine-into-human cfDNA mixtures was created from porcine and human plasma samples. Whole genome sequencing read counts mapping uniquely to porcine or human reference genomes were identified and counted to calculate the ratio of porcine cfDNA to total cfDNA in each sample. The correlation coefficient and accuracy (standard deviation; SD) was calculated.
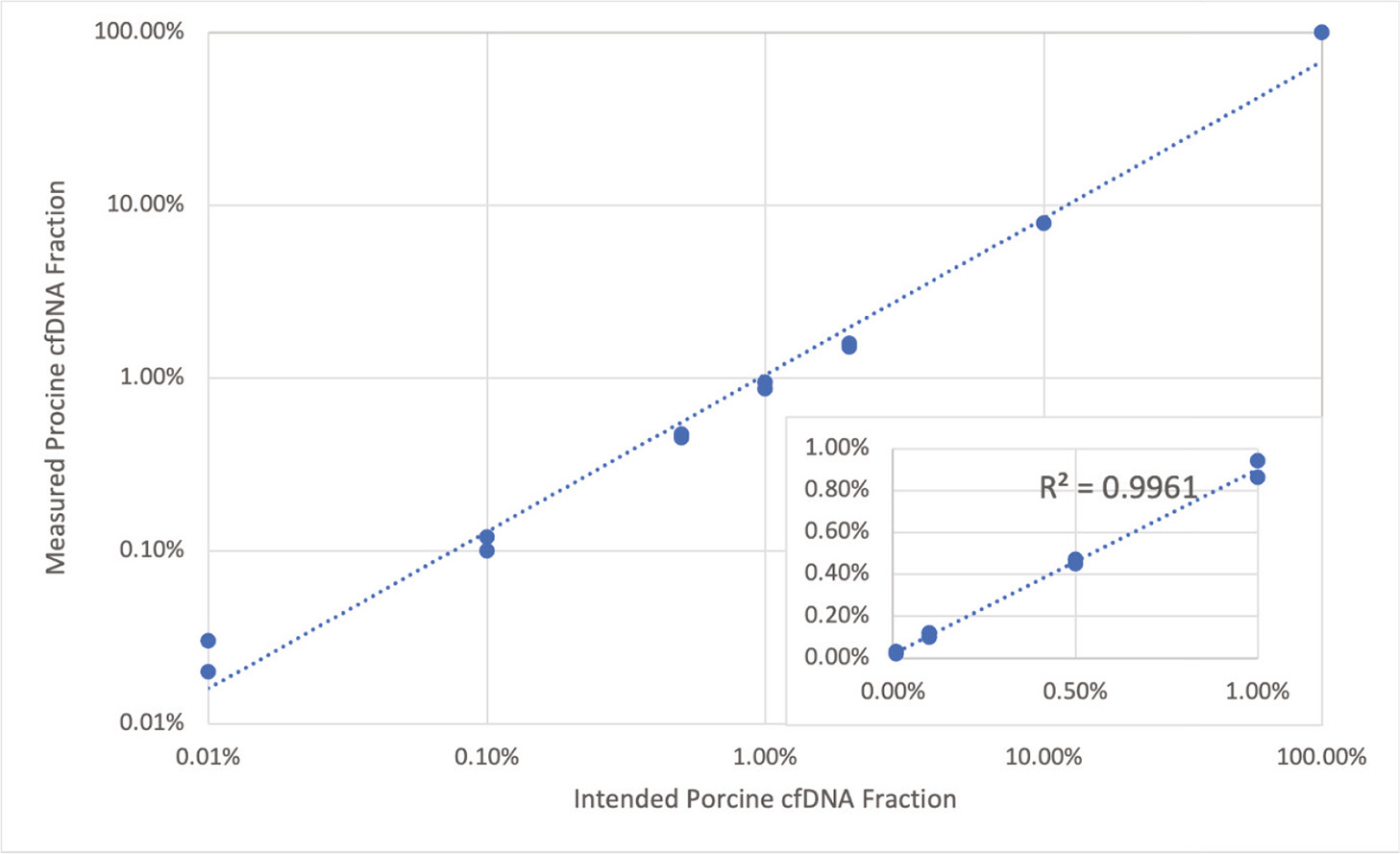
Results:The calculated porcine-derived cfDNA (pcfDNA) ratio in the 14 porcine-human cfDNA mixtures, ranging from 100% to 0.01% pcfDNA ratio, were plotted against the intended input pcfDNA ratio, yielding a concordance of r2= 0.9961 and SD of 0.029. At low and very low (<2% and <0.2%) pcfDNA values, SD remained low, at 0.025 and 0.011, respectively.
Conclusions: We demonstrated that whole genome sequencing can accurately detect and quantify porcine-derived cfDNA in mixtures with human cfDNA down to <0.1% pcfDNA fraction. This method is expected to support future research and clinical applications of solid organ xenotransplantation.
[1] Xenotransplants, Cell-free DNA, Donor-derived cell-free DNA, porcine cfDNA