
Sequential genome editing enhances triple-knockout efficiency and human transgene expression in porcine fibroblasts for xenotransplantation
Micaela Navarro1,2, Carolina Alvarez2, Bianca Estefania Devia2, Marcelo Perez-Pepe2, Yasemin Sezgin3, Genesis Snyder3, Adrián Abalovich 2, Meisam Naeimi Kararoudi3, Adrián Mutto1,2.
1Laboratorio de Biotecnologías Aplicadas a la Reproducción Animal, Instituto de Investigaciones Biotecnológicas “Dr. Rodolfo Ugalde”, Buenos Aires, Argentina; 2CrofaBiotech SA, Buenos Aires, Argentina; 3Center for Childhood Cancer and Blood Disease, Abigail Wexner Research Institute, Nationwide Children's Hospital, Columbus, OH, United States
Xenotransplantation using genetically modified pigs offers a promising alternative to solve the global organ shortage. Advances in gene editing have improved the safety and feasibility of pig-to-human transplants by overcoming key immunological and viral barriers. Notably, the triple knock-out (3-KO) of the main genes involved in hyperacute rejection -GGTA1, CMAH, and B4GalNT2- has significatively increased compatibility. In this study, we evaluated which is the most efficient strategy to generate 3-KO pig fibroblasts.
Dermal fibroblasts from PERV negative and mycoplasma-free pigs were edited using the CRISPR/Cas9 tool. Two gene-editing strategies were tested: all-at-once editing (AE) and sequential editing (SE). KOs were confirmed by sequencing, and protein expression was assessed by flow cytometry. Next, the 3-KO fibroblasts were transduced with lentiviruses encoding human genes involved in immune modulation and coagulation -CD39, CD46, CD47, CD55, and thrombomodulin (THBD)-. Successful insertions were validated by flow cytometry detecting human protein expression. Thereafter, edited fibroblasts were clonally isolated and used as donors for somatic cell nuclear transfer (SCNT). Finally, resulting blastocysts were transferred laparoscopically into synchronized surrogates, with ~150 embryos transferred/swine.
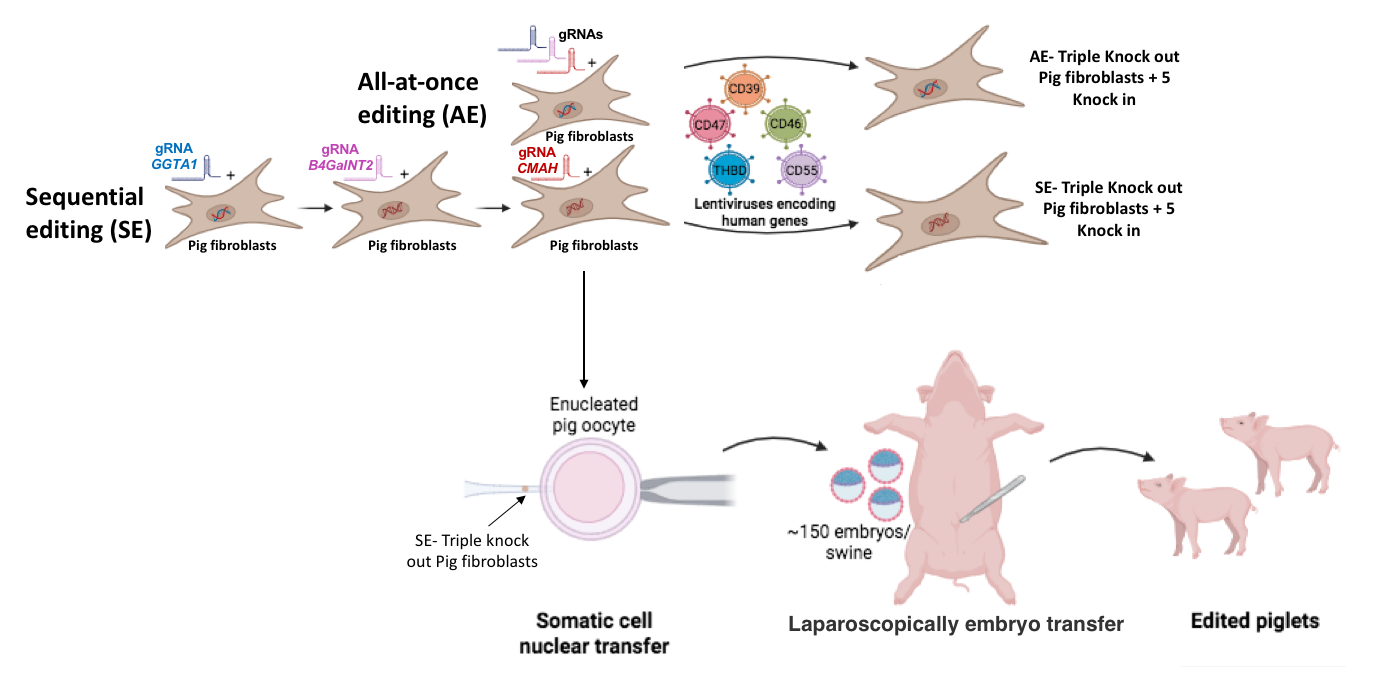
As results, ICE analysis revealed that the SE achieved higher KO-Scores for GGTA1, B4GalNT2, and CMAH (92%, 93%, and 87%, respectively) compared to AE (91%, 47%, and 50%, respectively). These findings were consistent with protein expression levels, where SE resulted in lower residual activity of all three proteins compared to the AE. Subsequently, 3-KO fibroblasts generated by SE or AE were transduced with lentiviral particles encoding the previously mentioned human genes. We observed that AE-derived 3-KO fibroblasts expressed lower levels of CD39, CD46, CD47, CD55, and THBD (39%, 40%, 77%, 74%, and 17%, respectively) than SE-derived cells (60%, 72%, 94%, 92%, and 49%, respectively).
Due to regulatory considerations, subsequent experiments were performed using the SE-derived 3-KO fibroblasts. Clonal isolation was performed, resulting in 12/30 clones, of which 8/12 carried heterozygous mutations and 4/12 displayed homozygous mutations. One of the homozygous clones was selected as a nuclear donor for SCNT and resulted in 17% of day 7- in vitro blastocyst development, compared to 30% when using wild-type donor cells (p<0.05, ANOVA). The resulting blastocysts were successfully implanted and established pregnancies, which are ongoing to date.
In summary, our findings demonstrate that sequential editing strategy is more efficient for generating porcine 3-KO fibroblasts compared to all-at-once. This approach also enhanced the expression of human transgenes and supported successful SCNT and embryo development. Our results represent a promising step toward the generation of genetically tailored porcine donors for xenotransplantation.
[1] Genetic edition
[2] Hyperacute rejection
[3] CRISPR/Cas9
[4] Somatic cell nuclear transfer
[5] Triple knock out
[6] Knock in of human genes
[7] Pig dermal fibroblasts
[8] Lentivirus
[9] GGTA1, CMAH, and B4GalNT2
[10] CD39, CD46, CD47, CD55, and thrombomodulin