Monitoring donor-derived cell free DNA in porcine heart xenotransplant recipients
Alesha Luxon3, Muhammad M Mohiuddin1, Bartley Griffith1, Avneesh Singh1, Sarah Cipriano1, David L Ayares2, Kimberly M Clark-Langone3, Dennis Fu3, Natalie Gulbahce3, Evgeniya Vaskova3, Robert N Woodward3, John Morlan3.
1Surgery, University of Maryland, Baltimore, MD, United States; 2Revivicor, Blacksburg, VA, United States; 3CareDx, Brisbane, CA, United States
Introduction: With recent advances in gene editing technology came the first heart transplants from genetically modified pigs into humans. While the recipients only survived for weeks post-transplant, similar technologies will undoubtably be used in future transplant patients. As xenotransplantation becomes more commonplace, the need for non-invasive diagnostic tools will also grow. Here we describe the development of 2 assays for monitoring donor derived cell-free DNA (dd-cfDNA) in the world’s first xenotransplant patients to receive a heart from a genetically modified pig.
Method: Blood was collected in Streck BCT tubes approximately weekly after transplant. cfDNA was extracted using the QIAamp Circulating Nucleic Acid Kit and quantitated by Qubit. Firstly, shotgun sequencing was performed on the Illumina NextSeq or MiSeq and data analyzed using an in-house bioinformatics pipeline. Secondly, digital PCR (dPCR) assays were designed to specifically detect exclusively either human or porcine sequences and genome copies of each species measured on the Stilla Naica system. In both cases, percentage of porcine dd-cfDNA was determined as well as recipient cfDNA.
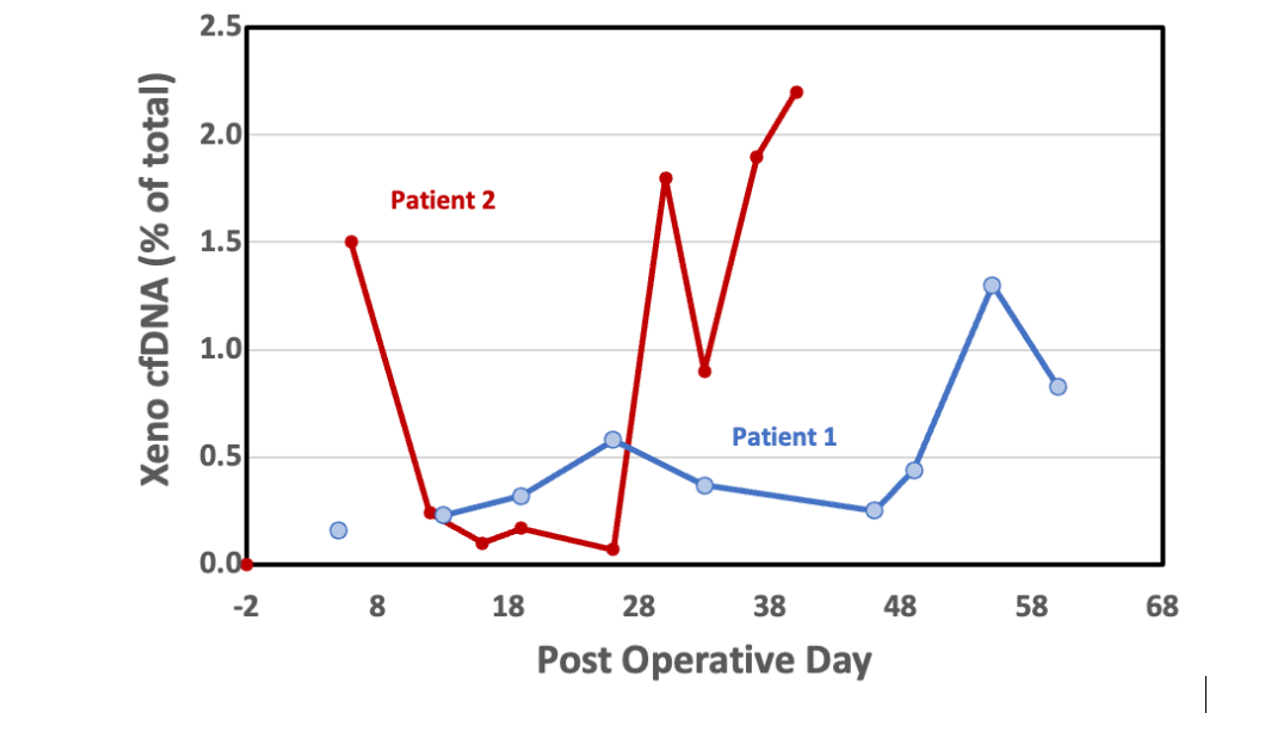
Results: Both cPCR and shotgun sequencing assays demonstrated practically zero dd-cfDNA signal in blank samples and showed good linearity with contrived samples. Patient dd-cfDNA results from shotgn and dPCR showed a vvery similar trend over time. dPCR results illustrated 0.12% dd-cfDNA at day 6 post-transplant rising to 0.79% by day 55 in Patient 1. Patient 2 had a high % dd-cfDNA initially that dropped but spiked again at the time of a major clinical event. This patient also had very high recipient cfDNA levels beginning at post-operative day 30.
Conclusion: Both shotgun sequencing and dPCR were successfully developed to monitor the world’s first Xenotransplant patients to receive a heart from a genetically modified pig. dPCR provides a cheap and rapid result, but accurate quantitation may be hindered by limited porcine genome copies. On the other hand, shotgun sequencing is more expensive but has the potential to reach lower limits of quantitation due to the larger number of targets being measured.
[1] monitoring
[2] dd-cfDNA
[3] donor derived cfDNA
[4] porcine derived cfDNA