
Endothelial injury and fibrogenesis drives post-transplant response after pig-to-human heart xenotransplantation
Amol Shetty1, Thomas Rousselle2, Haseeb Zubair2, Raphael Meier3, Daniel G Maluf3, Muhammad M Mohiuddin4, Valeria R Mas2.
1Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, MD, United States; 2Surgical Sciences Division, University of Maryland School of Medicine, Baltimore, MD, United States; 3Division of Transplantation, University of Maryland School of Medicine, Baltimore, MD, United States; 4Department of Surgery, University of Maryland School of Medicine, Baltimore, MD, United States
Introduction: The recent pig-to-human cardiac xenotransplants (XenoTx) performed at the University of Maryland Medical Center are at the forefront of continued significant advancements in the field of xenotransplantation. The cellular profiles of these XenoTx provide an avenue to better understand molecular perturbations needed to improve post-transplant (post-Tx) xenograft function. Evaluation of the cell-type specific expression profiles in context of the post-Tx clinical course and strategize ways to overcome the obstacle of antibody-mediated rejection (AMR) needed to advance the field of cardiac xenotransplantation.
Methods: We utilized post-Tx post-mortem wedge biopsies of the heart xenograft explant from each patient (Pt) for single-nuclei transcriptomic (snRNAseq) profiling using the 10X Genomics technology. We assessed quality control and generated raw gene count data using the CellRanger software. We integrated gene expression profiles for each Pt with snRNAseq profiles from native pig heart (nH) and analyzed the data using Seurat. Unsupervised cell clustering was performed using UMAP and cell-type specific molecular profiles were computed independently for each Pt.
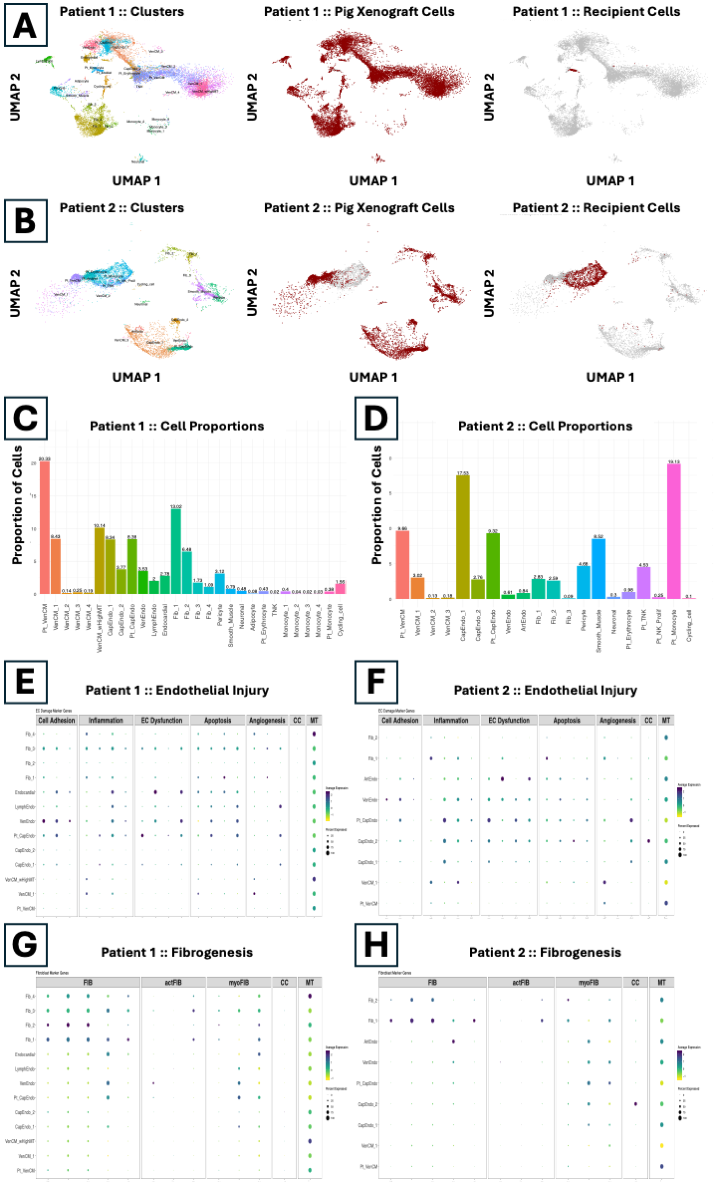
Results: A total of 58,700 high-quality nuclei (nH: 39,400; Pt 1: 12,600; Pt 2: 6,700) were analyzed and categorized into major cardiac cell-types, namely ventricular cardiomyocytes, endothelial, fibroblasts, smooth muscle, and neuronal (Fig 1A-B). We identified less than 1% of both resident pig and human immune cells in Pt 1 (Fig 1C), while Pt 2 showed about 24% of human immune cells (Fig 1D). Endothelial cells in both Pts showed overexpression of marker genes associated with cell adhesion, inflammation, endothelial dysfunction, and apoptosis indicating increased levels of endothelial injury (Fig 1E-F). Greater than 50% of fibroblasts from both Pts were PDGFRA+ and PDGFRB+ compared to fibroblasts from nH that were only PDGFRA+ (Fig 1G-H). In Pt 2, recipient-derived T and NK cells were enriched for genes associated with TCR signaling, NK-mediated cytotoxicity, and allograft rejection while recipient-derived monocytes were enriched for genes associated with chemokine signaling, neutrophil degranulation, and allograft rejection.
Conclusions: Overall, we identified heterogenous cellular compositions in the two XenoTx Pts with increased endothelial injury, increased fibrogenesis, and associated molecular profiles in both Pts. Immune profiles in Pt 2 also provided insights into early AMR and post-Tx immunosurveillance. Overall, our results provide clinicians a better understanding of genomic determinants of post-Tx xenograft function.
[1] Pig-to-Human Cardiac Xenotransplant
[2] Genomic Markers
[3] Endothelial Injury
[4] Fibrosis
[5] Immune Response